Background: The precise detection of cortical sleep spindles is critical to basic research on memory consolidationin rodents. Previous research using automatic spindle detection algorithms often lacks systematic parametervariations and validations.
New Method: We present a method to systematically tune and validate algorithm parameters in automaticspindle detection algorithms using a moderate number of human raters.
Results: Comparing a Hilbert transform-based algorithm to a ground truth constructed by six human raters, thismethod produced a parameter set yielding an F1 score of 0.82 at 10 ms resolution. The algorithm performancefell within the range of human agreement with the ground truth. Both human and algorithm failures aroselargely from disagreement in spindle boundaries rather than spindle occurrence. With no additional tuning, thealgorithm performed similarly in recordings from different days or rats.
Comparison with existing methods: Most spindle detection algorithms do not perform systematic parameter var-iations and validation using a ground truth. To our knowledge, our study is thefirst in which rodent spindle datais scored by humans, and in which an automatic spindle detection algorithm is evaluated with respect to thisground truth. The rodent data from this study make it possible to compare our algorithm with others previouslytested on human data.
Conclusions: We present a general ground truth based approach for the tuning and validation of spindle ex-traction algorithms and suggest that algorithms aimed at extracting precise spindle timing in rats should use asystematic approach for parameter tuning.
Full paper can be found here. The code to reproduce the figures can be found below.
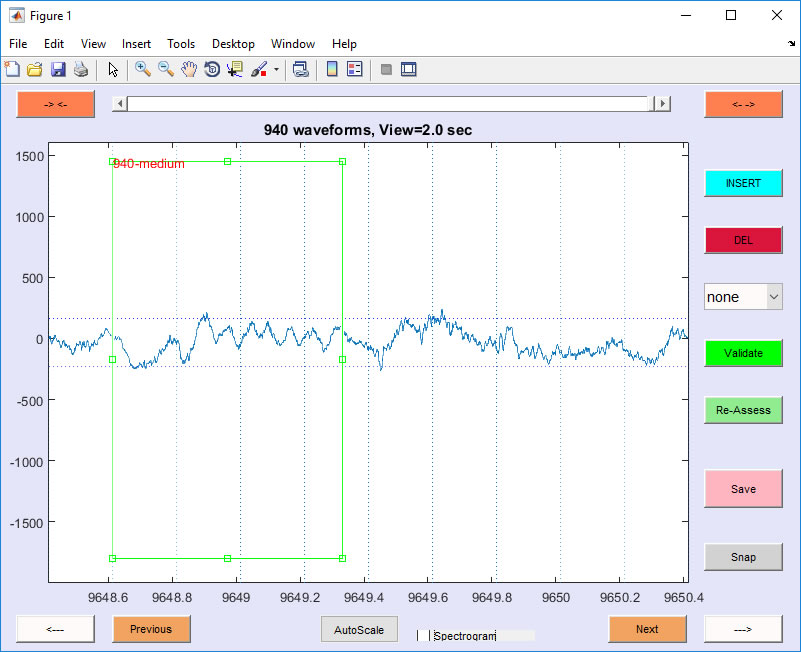
This code is general and allows the user to read in one or more EEG files (Neuralynx CSC format, recorded simultaneously), to scroll though the traces and place bounding boxes around graphoelements of interest. It also allows for user-defined scoring of each graphoelement (e.g. 'good', 'medium', 'bad'). Results are saved in a matlab file that can be appended/edited later. 
Example (Spindle scoring): This example uses a raw datafile CSC5.ncs. All datafiles can be downloaded. The code can be downloaded here. Create a folder CENL and place the unziped downloaded files Data and Code it. In Matlab type these two lines (replace the '...' with the absolute path to the CENL folder you created):
addpath(genpath('.....\CENL\Code'));
RootFolder='...\CENL';
A brief explanation of the scoring scheme can be found here. Follow the steps below to start the EEGscoring interface (in this case for Spindles).
RawData=[RootFolder '\Data\SpindleData\CSC5.ncs'];
SPDLFile=[RootFolder '\Data\SpindleData\GroundTruth\Eliot020517Recording-Rater3-SPDLs.mat']; %optional. A file that contains the boundingboxes of some of the spindles. This one contains all the spindles extracted by user 'Rater3' (we include 5 other users in this folder). If you load such a file, then rateEEG() will update it...
data=load(SPDLFile);
stops=data.Stops; % (optional) array of times when the rat is considered immobile (according to the EMG)
tstart=650209091; tend=9651407092; % time range to work on (micro-seconds)
RateEEG(SPDLFile,RawData,tstart,tend, stops,{'good','medium','bad','k-less','k-alone'});
Rate EEG will startup and ask if you want to load or overwrite the SPDLFile. Choose 'Load'. This file contains 940 manually scored spindles. Click 'Previous' to jump to the previously scored spindles (type help rateEEG to see more information).
If you modify/improve this utility, please let us know and we will link your code here!
- Figure 2
- Figure 3
- Figure 4
- Figure 5 and 6
- Figure 7
Brought to you by the Computational and Experimental Neuroscience Laboratory